
Figure 1. Residues collected from inside an Old Edgefield jar from a private collection. The appearance was highly varied ranging from mixtures of viscous liquids with solid particles to dry solid flakes.
As part of the exhibition, Hear Me Now: The Black Potters of Old Edgefield, South Carolina, The Met’s Department of Scientific Research (DSR) investigated organic food residues found inside large nineteenth-century alkaline-glazed stoneware vessels from the Old Edgefield District, South Carolina. “Examining Storage Jars from the American South” describes the driving questions about the jars’ use and the users’ lifestyle. Investigations reported in “The Inside (and Outside) Scoop: Scientific Analysis of Food Residues Inside the Jars from Old Edgefield, South Carolina” established that the heterogeneous residues are mostly oily materials with solid materials of various unknown origins. We hoped to gain more information about the jars’ contents from these residues, but to do so we need the sophisticated tools and expertise of our collaborators through ARCHE.
Samples from cultural heritage, including these residues, are typically a complex mixture of materials used to make the object, or in this case how it was used, but also the materials accumulated through use and time (grime, insects, degradation products of the original or deposited materials, etc.). Light-based techniques such as Fourier transform infrared spectroscopy (FTIR) can only tell us the class of materials present (lipid, wax, protein). Separation techniques coupled to mass spectrometry, such as Evolved Gas Analysis-Mass Spectrometry (EGA-MS) and Thermally assisted Hydrolysis and Methylation Pyrolysis Gas-Chromatography-Mass Spectrometry (THM-Py-GCMS), can provide more specific molecular identification. Where the material is a fat or oil, as in our case, this includes saturated and unsaturated fatty acids and their degradation products. However, these techniques are limited to small molecules. We require much more powerful mass spectrometry techniques to tap into the tissue and species-specific information trapped in the subtle variations of much larger biomolecules, such as triglycerides or proteins. Our department accesses these techniques through our ARCHE (ARt and Cultural HEritage: Natural Organic Polymers by Mass Spectrometry) collaboration with the University of Bordeaux in France.
Proteomics and lipidomics techniques are commonly associated with biological or medical research. These powerful tools are designed to look at all the proteins or lipids in a biological system, such as a plant seed or a tumor cell. They are extremely sensitive techniques and very accurate. Within a sample, tiny amounts of materials can be unequivocally identified as a specific protein belonging to a particular animal species, sometimes even a specific tissue, or specific lipids characteristic of a specific tissue. This time intensive research requires complex instrumentation, expert knowledge, and curated databases.

Figure 2. Vaclav Krupicka (University of Bordeaux) preparing a sample for injection onto the ThermoFisher Orbitrap Eclipse Tribrid Mass Spectrometer. Credit: Vaclav Krupicka and Daniel Vallejo (Georgia Institute of Technology)
Cultural heritage objects such as Edgefield pottery can exploit these powerful techniques because organic residues, as well as early paint binders, glues/adhesives, ivory, bone, shell, horn, textiles, and coatings, all originate from an animal or plant source. Think of an artwork as its own biological system of materials. Since historians assign the jars as vessels for preserved meat (muscle tissue), which is predominantly made of proteins, our first approach was to look for proteins in the residues. Even though the residues are almost exclusively lipidic, and the jars had been rinsed with hot water, there is still a chance that there are trace amounts of proteins trapped in the oily matrix. Another favorable factor is that the proteins for muscle tissue for many of our suspect species (cow, deer, pig) are already in public databases.
Proteins are strings of amino acids, which, when folded properly, can provide structure to cells or perform activities to keep cells alive. The sequence of the amino acids in the string are written by the DNA sequence in genes. Each species has unique DNA sequences, which can translate into unique amino sequences of its proteins. For proteins such as collagens (skin, bone) and casein (milk), much of the proteins’ amino acid sequence will be conserved. Still, there can be enough unique changes between species that allow the discrimination of species source, in our case: cow, deer, or pig.
For this investigation, we employ bottom-up proteomics. This approach uses enzymes to break proteins into predictable smaller pieces called peptides. These peptides are then separated and fragmented so their sequences can be reconstructed, and the proteins and possible species sources can be identified by searching against existing databases.
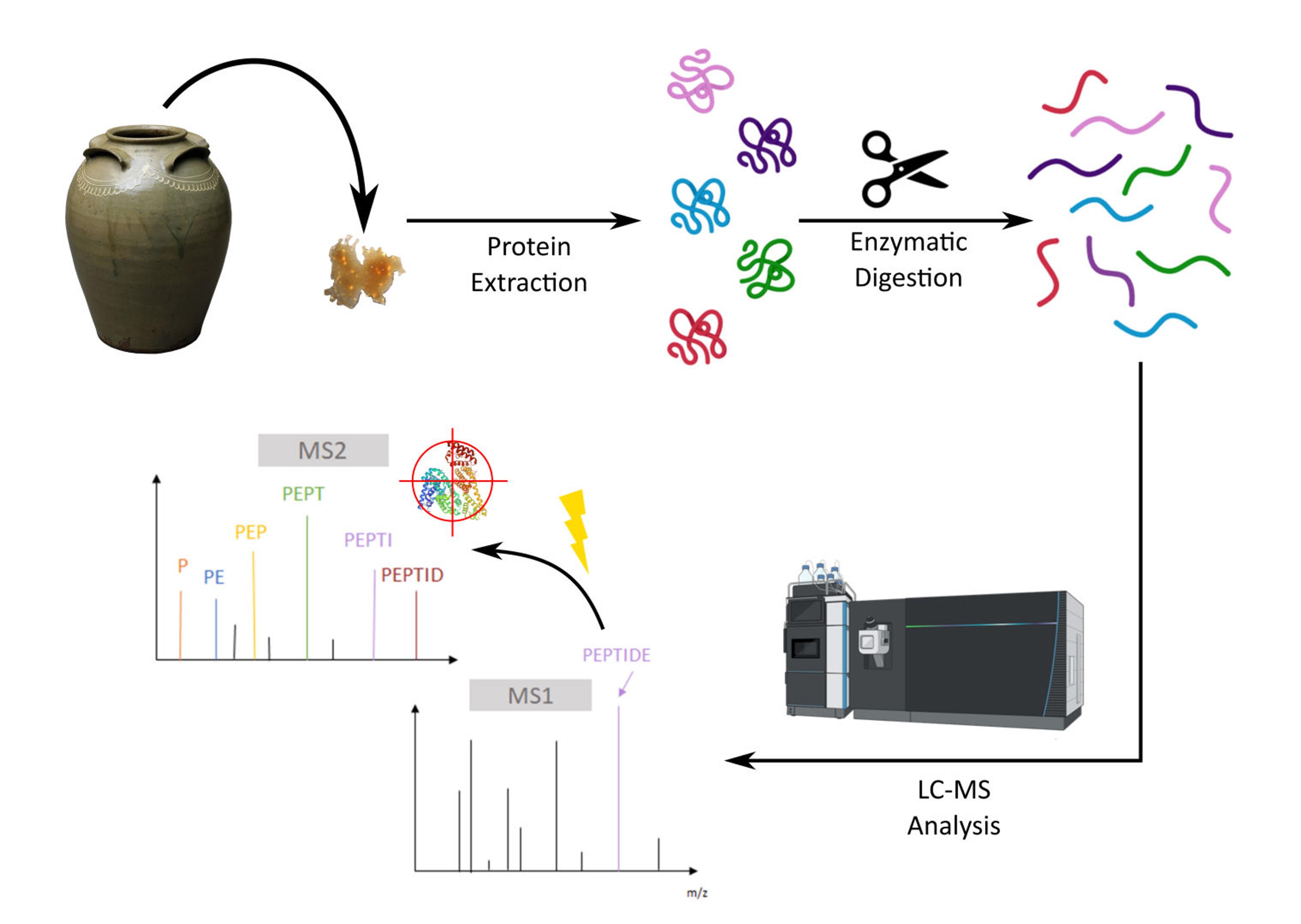
Figure 3. Diagram showing bottom-up MS proteomics workflow including MS and MS2 steps that allow for sequencing of observed peptides and subsequent identification of proteins present in the sample. Credit: Vaclav Krupicka (University of Bordeaux).
The oily residues held their secrets well. Residues from three jars were analyzed, one showing trace amounts of proteins by FTIR. In all the samples, the keratins and collagens, the proteins found in hair and skin, were discovered using proteomics. These are most associated with contamination. The analysis did not reveal animal muscle tissue proteins such as myosins or actins in two of the samples. However, in the third sample, which had traces of protein by FTIR, animal muscle tissue proteins were identified, surprisingly, not for pigs or cows but for insects!
While this result is disappointing in terms of identifying the jars’ contents, it demonstrates how sensitive and specific proteomics can be. Proteomics has femtomolar sensitivity, and the ARCHE laboratory can achieve hundreds of attomoles and less with particular settings for the instrument. Since we detect muscle proteins from insect detritus, we surely would discern muscle proteins from preserved meat, if they were present. While we could continue to analyze the residues from the remaining three jars for proteins, lipid analysis may be more promising. Since the sample has more material available, although lipids hold less genetic information, they are often still characteristic of the tissue and could be distinct between species and provide clearer molecular indicators of food preservation practices.
Lipids would occur in a rendered fat like lard or the intramuscular fat found within muscle tissue (meat) that could have been stored in the jars. In general, lipids are a broad group of molecules that include the triacylglycerols found in adipose tissue (fat) and plant fats. Lipidomics interrogates this variety of molecules to differentiate between animals and plants but also different species of each. However, the databases for lipids are not as well developed as the protein databases for proteomics. Therefore, a significant investment into the creation of lipid databases for the species of interest (cow, pig, deer) and the preservation method (pickling, salting, sugaring, heating) is needed. Collaboration with food historians knowledgeable in the diet and preservation methods of nineteenth-century Edgefield, South Carolina, can guide the formation of a reference library of materials to be prepared and tested.
